科技工作者之家
科技工作者之家APP是专注科技人才,知识分享与人才交流的服务平台。
科技工作者之家 2019-02-21

欢迎收听本周由Shamini Bundell和Nick Howe带来的一周科学故事,本期播客片段讨论通过虚拟筛选数百万种化学物质,加速药物发现。欢迎前往iTunes或你喜欢的其他播客平台下载完整版,随时随地收听一周科研新鲜事。

音频文本:
Interviewer: Nick Howe
Discovering a new medical drug is a long and resource-intensive process. It can take a decade or more for a drug from the drawing board to the hospital ward, in a process that involves a huge number of researchers. But for every drug that makes it, many more fall by the wayside in a process that costs billions of dollars. Naturally, researchers are keen to streamline and accelerate the discovery process in order to save money and time. One possible way to do this is by using computer simulations to screen early drug candidates. Whilst this technology has existed for some time, this week in Nature, there’s a paper describing how size is important in the world of virtual screening, as Bryan Roth from the University of North Carolina, one of the co-authors of the paper, describes.
Interviewee: Bryan Roth
We were trying to basically see if we could enhance the process by several orders of magnitude, by speeding up the initial screening process. The initial screen typically would take months, and is usually limited to, say, 100,000 compounds to maybe 1 million compounds.
Interviewer: Nick Howe
Usually the initial screening involves physically testing thousands of compounds. In this case, rather than testing the activity of a relatively small number of compounds against a particular drug target in the lab, the team used a ‘virtual library’ – a catalogue of hundreds of millions of compounds.
Interviewee: Bryan Roth
And so, what we did here was we took advantage of a large resource of potential chemicals – these are chemicals that are theoretical in that they have not yet been synthesised but could be made by chemists using sort of common chemical reactions or typical chemical reactions, and this is using a resource called a zinc database, which was developed by my co-authors Brian Shoichet and John Irwin at UCSF. And over the last few years, they’ve been able to increase the size of the zinc database to nearly 1 billion compounds, so a large number of compounds, so more compounds than one could ever physically screen.
Interviewer: Nick Howe
By using a lot of computing power, the team were able to virtually test the interactions of these compounds against a number of drug targets in a process known as ‘automated docking’. You might think that simulating more automated docking would slow down the discovery process, but Bryan suggests that for a number of reasons, when it comes to virtual libraries, bigger is better.
Interviewee: Bryan Roth
The main advantage of a large library is you get access to a large number of different chemotypes, so these different molecules with different shapes. And it’s become clear over the past several years that if you have compounds that have unique shapes, then when they interact with the target they’re likely to have interesting biological activities that never would have been predicted before and ultimately could be therapeutic in ways that were previously unanticipated.
Interviewer: Nick Howe
By testing such a diverse array of chemical structures, including molecular shapes that are completely novel, the team were able to find interesting chemical reactions that haven’t been seen before, and they were able to do it quickly. For example, in just over a day, the researchers were able to virtually test over 138 million compounds for their ability to dock with the D4 dopamine receptor, a drug target for treatment of diseases such as Parkinson’s. The team also looked for inhibitors of a bacterial enzyme called AmpC β-lactamase, involved in antibiotic resistance. By screening 99 million molecules, they found some promising candidates. Out of these, they synthesised a very potent chemical that appears to have a completely new mechanism of action. And this is the proposed advantage of this technique – going from rapidly screening millions of compounds in the computer to then choosing a top list to synthesise and test in the lab. This approach could massively cut down on the time needed to identify early drug candidates.
Interviewee: Bryan Roth
It turns out that having this huge library really is a tremendous resource basically because you get access to this chemical space that there is no way to physically ever obtain, and it turns out that that is a huge, huge advantage, one that we had not anticipated. We hoped that we would get that, but we didn’t really anticipate just how valuable it was.
Interviewer: Nick Howe
Of course, there’s a long way from discovering a chemical interaction to developing a drug for clinical use, but the time saved could be incredibly valuable. David Gloriam from the University of Copenhagen has written a News and Views article on this topic. He thinks that although virtual screening has been around for a while, there’s a lot to appreciate in this new work.
Interviewee: David Gloriam
Well, the method of virtual screening has been refined and applied for two or three decades now, but what is really setting their approach apart is the huge compound libraries that they screen, and previously this has not been possible because of computational limitations and maybe for many groups because of licensing issues to do with the software, and also the availability of these compound databases.
Interviewer: Nick Howe
Like with any developing technique, there are some caveats that need to be considered. For docking to work, you need to have well modelled structures of targets and even then, it will only work on molecules that contain particular binding sites that allow for the activity to be changed. David thinks that there are some other challenges of a less scientific nature too. As drug discovery is an expensive process, companies often seek unique compounds that they can patent and have exclusive rights to produce. The massive library in this paper, however, is open for anyone to use.
Interviewee: David Gloriam
Because many of these molecules are available open access, then is the question of whether this would restrict rights to having invented this molecule. There are indications that if you make compounds available on the internet, this could limit the possibilities to make a stronger type of patent.
Interviewer: Nick Howe
If the structures are freely available, then the question is whether you can claim to have invented the molecule, which is a requirement for the stronger type of patent. Without the security of a powerful patent it’s reasonable to ask whether drug companies would want to invest in drug discovery like this. Both David and Bryan are confident that these challenges could be overcome. Also, as more drug targets are getting better described, this method could be applied more widely in the future. Right now, we can already start using this approach to look for novel drugs with the online ZINC15 database. You can read Bryan’s paper along with David’s News and Views article over at nature.com/nature.ⓝ
Nature Podcast每周为您带来科学世界的全球新闻故事,覆盖众多科研领域,重点讲述Nature期刊上激动人心的研究故事。我们将话筒递给研究背后的科学家,呈现来自Nature记者和编辑的深度分析。在2017年,来自中国的收听和下载超过50万次,居全球第二。
↓↓iPhone用户长按二维码进入iTunes订阅

↓↓安卓用户长按二维码进入推荐平台acast订阅

点击“阅读原文”访问Nature官网收听完整版播客

来源:Nature-Research Nature自然科研
原文链接:https://mp.weixin.qq.com/s?__biz=MzAwNTAyMDY0MQ==&mid=2652558589&idx=2&sn=6f3759795f3afbc0ccc84b6801d4d250&scene=0#wechat_redirect
版权声明:除非特别注明,本站所载内容来源于互联网、微信公众号等公开渠道,不代表本站观点,仅供参考、交流、公益传播之目的。转载的稿件版权归原作者或机构所有,如有侵权,请联系删除。
电话:(010)86409582
邮箱:kejie@scimall.org.cn

光催化,今日《Nature》!
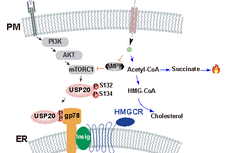
武汉大学今日Nature;华大基因Nature

重返月球 | Nature Podcast

“复苏”大脑| Nature Podcast

合成“胚胎” | Nature Podcast

这篇《Nature》会发光!
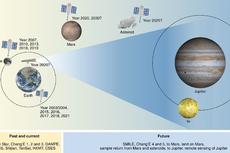
魏勇等-Nature Astronomy:中国的行星探索路线图

蒲慕明院士:建设世界一流期刊关系学术创新的发言权
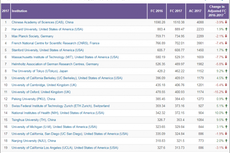
中科院位列2018自然指数榜单全球十大科研机构首位

仿生,登上Nature封面!